Attributes of Animals
animals.RdThis data set considers 6 binary attributes for 20 animals.
data(animals)
Format
A data frame with 20 observations on 6 variables:
| [ , 1] | war | warm-blooded |
| [ , 2] | fly | can fly |
| [ , 3] | ver | vertebrate |
| [ , 4] | end | endangered |
| [ , 5] | gro | live in groups |
| [ , 6] | hai | have hair |
All variables are encoded as 1 = 'no', 2 = 'yes'.
Source
Leonard Kaufman and Peter J. Rousseeuw (1990): Finding Groups in Data (pp 297ff). New York: Wiley.
Details
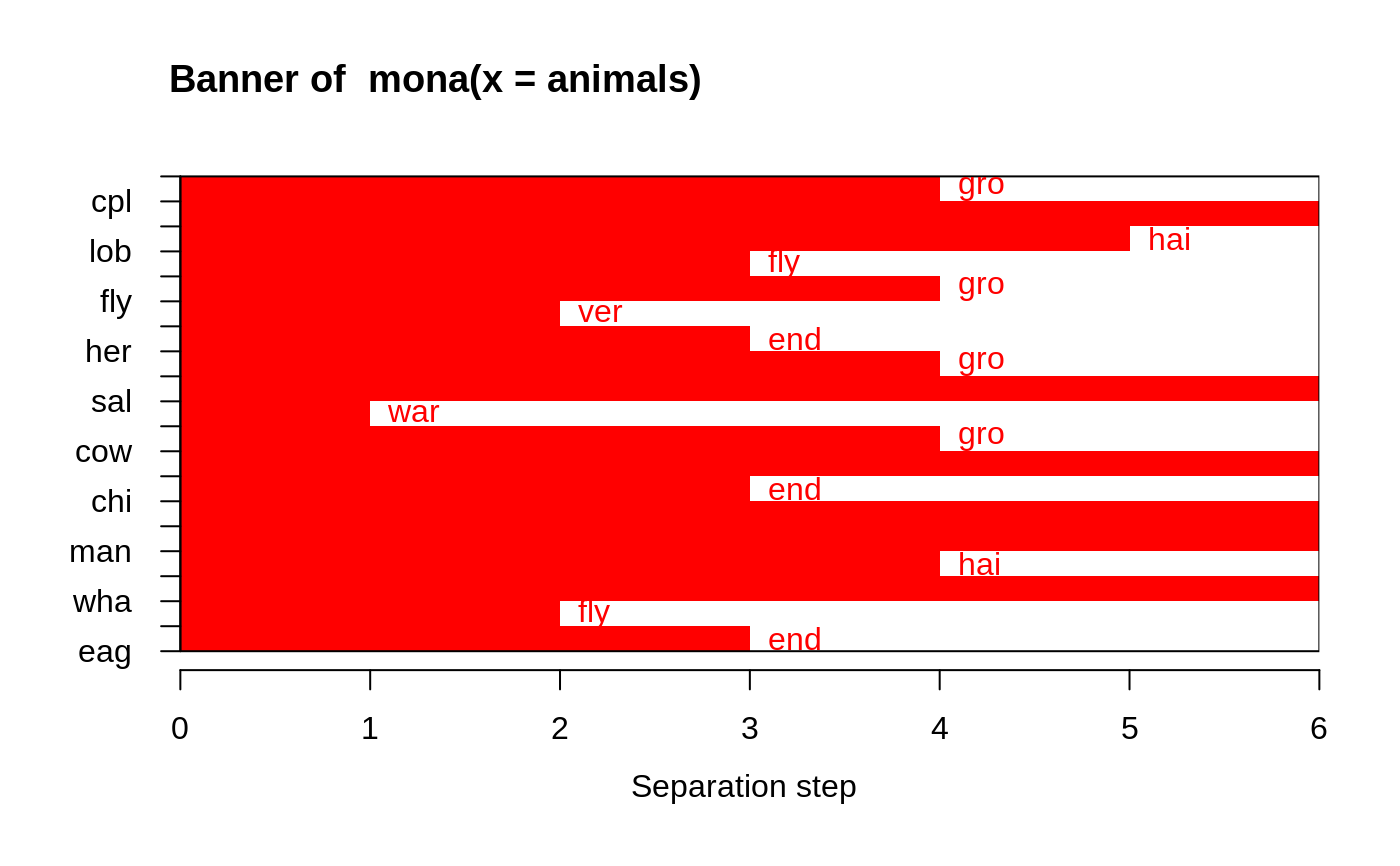
This dataset is useful for illustrating monothetic (only a single variable is used for each split) hierarchical clustering.
References
see Struyf, Hubert & Rousseeuw (1996), in agnes.
Examples
#> war fly ver end gro hai #> 1 10 16 6 12 6 11 #> 2 10 4 14 6 11 9#> mona(x, ..) fit; x of dimension 20x6 #> Because of NA's, revised data: #> war fly ver end gro hai #> ant 0 0 0 0 1 0 #> bee 0 1 0 0 1 1 #> cat 1 0 1 0 0 1 #> cpl 0 0 0 0 0 1 #> chi 1 0 1 1 1 1 #> cow 1 0 1 0 1 1 #> duc 1 1 1 0 1 0 #> eag 1 1 1 1 0 0 #> ele 1 0 1 1 1 0 #> fly 0 1 0 0 0 0 #> fro 0 0 1 1 0 0 #> her 0 0 1 0 1 0 #> lio 1 0 1 1 1 1 #> liz 0 0 1 0 0 0 #> lob 0 0 0 0 0 0 #> man 1 0 1 1 1 1 #> rab 1 0 1 0 1 1 #> sal 0 0 1 0 0 0 #> spi 0 0 0 0 0 1 #> wha 1 0 1 1 1 0 #> Order of objects: #> [1] ant cpl spi lob bee fly fro her liz sal cat cow rab chi lio man ele wha duc #> [20] eag #> Variable used: #> [1] gro NULL hai fly gro ver end gro NULL war gro NULL end NULL NULL #> [16] hai NULL fly end #> Separation step: #> [1] 4 0 5 3 4 2 3 4 0 1 4 0 3 0 0 4 0 2 3 #> #> Available components: #> [1] "data" "hasNA" "order" "variable" "step" "order.lab" #> [7] "call"