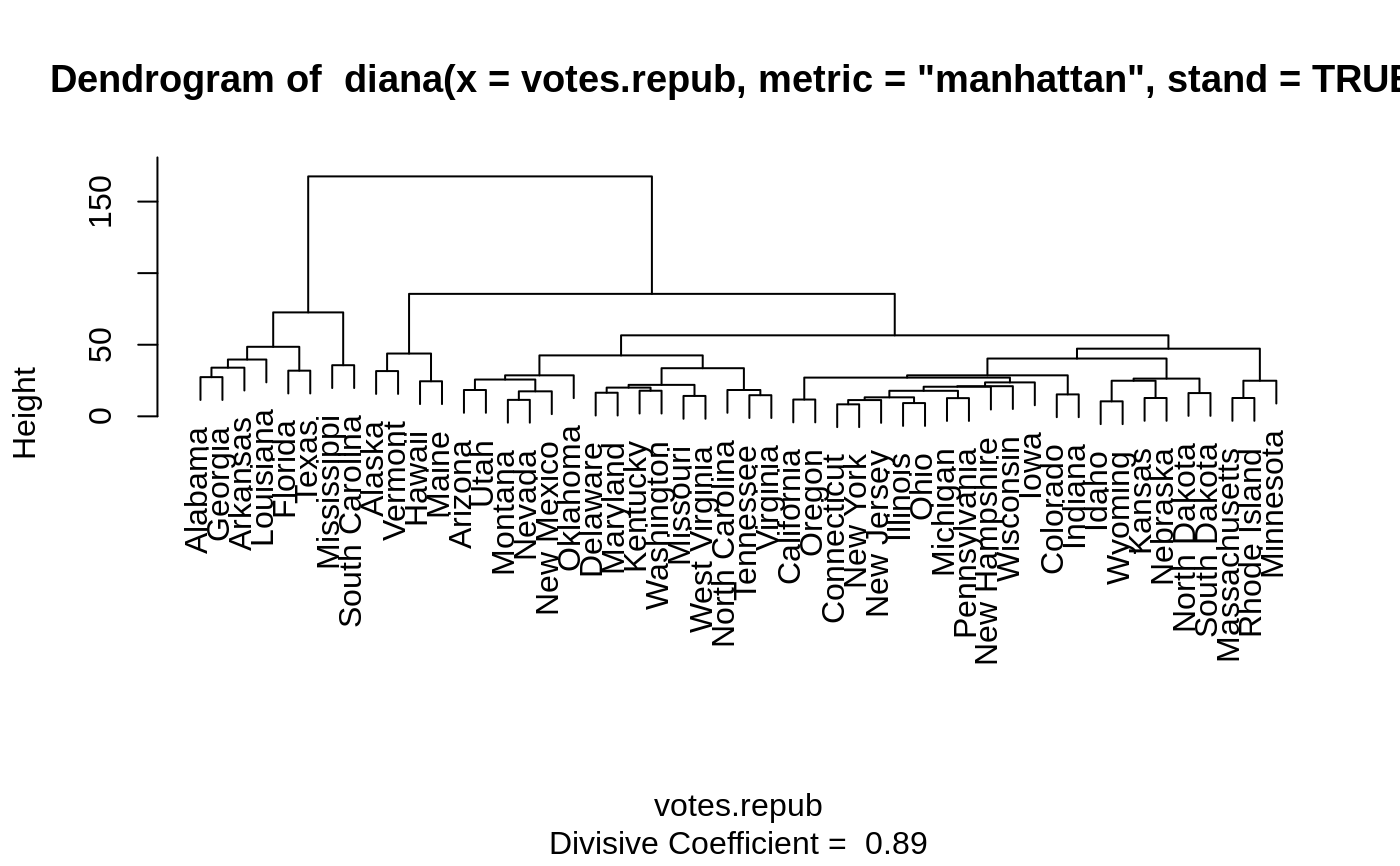
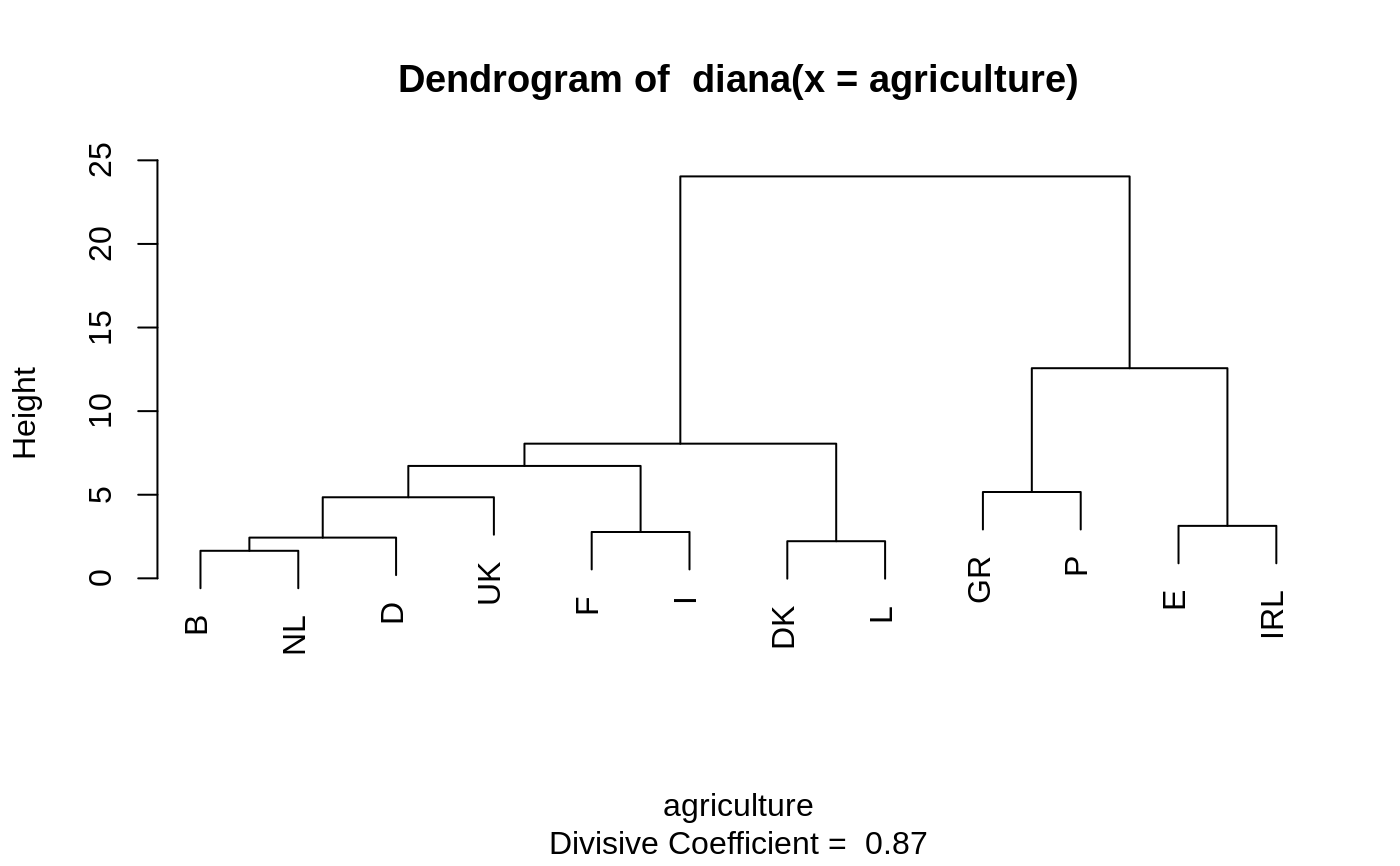
DIvisive ANAlysis Clustering
diana.RdComputes a divisive hierarchical clustering of the dataset
returning an object of class diana.
diana(x, diss = inherits(x, "dist"), metric = "euclidean", stand = FALSE, stop.at.k = FALSE, keep.diss = n < 100, keep.data = !diss, trace.lev = 0)
Arguments
| x | data matrix or data frame, or dissimilarity matrix or object,
depending on the value of the In case of a matrix or data frame, each row corresponds to an observation,
and each column corresponds to a variable. All variables must be numeric.
Missing values ( In case of a dissimilarity matrix, |
|---|---|
| diss | logical flag: if TRUE (default for |
| metric | character string specifying the metric to be used for calculating
dissimilarities between observations. |
| stand | logical; if true, the measurements in |
| stop.at.k | logical or integer, |
| keep.diss, keep.data | logicals indicating if the dissimilarities
and/or input data |
| trace.lev | integer specifying a trace level for printing
diagnostics during the algorithm. Default |
Value
an object of class "diana" representing the clustering;
this class has methods for the following generic functions:
print, summary, plot.
Further, the class "diana" inherits from
"twins". Therefore, the generic function pltree can be
used on a diana object, and as.hclust and
as.dendrogram methods are available.
A legitimate diana object is a list with the following components:
a vector giving a permutation of the original observations to allow for plotting, in the sense that the branches of a clustering tree will not cross.
a vector similar to order, but containing observation labels
instead of observation numbers. This component is only available if
the original observations were labelled.
a vector with the diameters of the clusters prior to splitting.
the divisive coefficient, measuring the clustering structure of the
dataset. For each observation i, denote by \(d(i)\) the diameter of the
last cluster to which it belongs (before being split off as a single
observation), divided by the diameter of the whole dataset. The
dc is the average of all \(1 - d(i)\). It can also be seen
as the average width (or the percentage filled) of the banner plot.
Because dc grows with the number of observations, this
measure should not be used to compare datasets of very different
sizes.
an (n-1) by 2 matrix, where n is the number of
observations. Row i of merge describes the split at step n-i of
the clustering. If a number \(j\) in row r is negative, then the single
observation \(|j|\) is split off at stage n-r. If j is positive, then the
cluster that will be splitted at stage n-j (described by row j), is
split off at stage n-r.
an object of class "dissimilarity", representing the total
dissimilarity matrix of the dataset.
a matrix containing the original or standardized measurements, depending
on the stand option of the function agnes. If a
dissimilarity matrix was given as input structure, then this component
is not available.
Details
diana is fully described in chapter 6 of Kaufman and Rousseeuw (1990).
It is probably unique in computing a divisive hierarchy, whereas most
other software for hierarchical clustering is agglomerative.
Moreover, diana provides (a) the divisive coefficient
(see diana.object) which measures the amount of clustering structure
found; and (b) the banner, a novel graphical display
(see plot.diana).
The diana-algorithm constructs a hierarchy of clusterings,
starting with one large
cluster containing all n observations. Clusters are divided until each cluster
contains only a single observation.
At each stage, the cluster with the largest diameter is selected.
(The diameter of a cluster is the largest dissimilarity between any
two of its observations.)
To divide the selected cluster, the algorithm first looks for its most
disparate observation (i.e., which has the largest average dissimilarity to the
other observations of the selected cluster). This observation initiates the
"splinter group". In subsequent steps, the algorithm reassigns observations
that are closer to the "splinter group" than to the "old party". The result
is a division of the selected cluster into two new clusters.
See also
agnes also for background and references;
cutree (and as.hclust) for grouping
extraction; daisy, dist,
plot.diana, twins.object.
Examples
#> Merge: #> [,1] [,2] #> [1,] -7 -32 #> [2,] -13 -35 #> [3,] -12 -50 #> [4,] 1 -30 #> [5,] -26 -28 #> [6,] -5 -37 #> [7,] -22 -38 #> [8,] -21 -39 #> [9,] -16 -27 #> [10,] 4 2 #> [11,] -25 -48 #> [12,] -42 -46 #> [13,] -6 -14 #> [14,] -34 -41 #> [15,] -8 -20 #> [16,] 5 -31 #> [17,] 10 7 #> [18,] -17 -47 #> [19,] -3 -44 #> [20,] -33 12 #> [21,] 15 18 #> [22,] 17 -29 #> [23,] 22 -49 #> [24,] 21 11 #> [25,] 23 -15 #> [26,] -11 -19 #> [27,] 3 9 #> [28,] 8 -23 #> [29,] 19 16 #> [30,] 27 14 #> [31,] 6 25 #> [32,] -1 -10 #> [33,] 31 13 #> [34,] 29 -36 #> [35,] -2 -45 #> [36,] -9 -43 #> [37,] 24 20 #> [38,] 32 -4 #> [39,] -24 -40 #> [40,] 38 -18 #> [41,] 33 30 #> [42,] 34 37 #> [43,] 35 26 #> [44,] 41 28 #> [45,] 40 36 #> [46,] 42 44 #> [47,] 45 39 #> [48,] 43 46 #> [49,] 47 48 #> Order of objects: #> [1] Alabama Georgia Arkansas Louisiana Florida #> [6] Texas Mississippi South Carolina Alaska Vermont #> [11] Hawaii Maine Arizona Utah Montana #> [16] Nevada New Mexico Oklahoma Delaware Maryland #> [21] Kentucky Washington Missouri West Virginia North Carolina #> [26] Tennessee Virginia California Oregon Connecticut #> [31] New York New Jersey Illinois Ohio Michigan #> [36] Pennsylvania New Hampshire Wisconsin Iowa Colorado #> [41] Indiana Idaho Wyoming Kansas Nebraska #> [46] North Dakota South Dakota Massachusetts Rhode Island Minnesota #> Height: #> [1] 27.363453 33.969252 39.658259 48.534276 31.899654 72.598496 #> [7] 35.691518 167.580197 31.582223 43.846009 24.487963 85.552482 #> [13] 18.393392 25.676314 11.493967 17.455521 28.625502 42.544800 #> [19] 16.485096 20.044499 17.875161 21.983729 14.218077 33.610713 #> [25] 18.397326 14.757619 56.556754 11.701321 27.058874 8.382005 #> [31] 11.368197 13.252375 9.230040 17.834836 12.708189 20.667139 #> [37] 21.039972 23.665856 28.605405 15.317027 40.339045 10.462936 #> [43] 24.835249 12.804188 26.362915 16.251922 47.257725 12.791603 #> [49] 24.872061 #> Divisive coefficient: #> [1] 0.8869182 #> #> Available components: #> [1] "order" "height" "dc" "merge" "diss" "call" #> [7] "order.lab" "data"plot(dv)#> dv2 #> 1 2 #> 8 42#> [1] "Alabama" "Arkansas" "Florida" "Georgia" #> [5] "Louisiana" "Mississippi" "South Carolina" "Texas"## For two groups, does the metric matter ? dv0 <- diana(votes.repub, stand = TRUE) # default: Euclidean dv.2 <- cutree(as.hclust(dv0), k = 2) table(dv2 == dv.2)## identical group assignments#> #> TRUE #> 50#> --[dendrogram w/ 2 branches and 50 members at h = 31.1] #> |--[dendrogram w/ 2 branches and 8 members at h = 15.3] #> | |--[dendrogram w/ 2 branches and 6 members at h = 10.5] #> | | |--[dendrogram w/ 2 branches and 3 members at h = 8.46] #> | | | |--[dendrogram w/ 2 branches and 2 members at h = 6.28] #> | | | | |--leaf "Alabama" #> | | | | `--leaf "Georgia" #> | | | `--leaf "Louisiana" #> | | `--[dendrogram w/ 2 branches and 3 members at h = 7.68] #> | | |--[dendrogram w/ 2 branches and 2 members at h = 7.1] #> | | | |--leaf "Arkansas" #> | | | `--leaf "Florida" #> | | `--leaf "Texas" #> | `--[dendrogram w/ 2 branches and 2 members at h = 9.55] #> | |--leaf "Mississippi" #> | `--leaf "South Carolina" #> `--[dendrogram w/ 2 branches and 42 members at h = 17] #> |--[dendrogram w/ 2 branches and 4 members at h = 9.43] #> | |--[dendrogram w/ 2 branches and 2 members at h = 6.45] #> | | |--leaf "Alaska" #> | | `--leaf "Vermont" #> | `--[dendrogram w/ 2 branches and 2 members at h = 5.23] #> | |--leaf "Hawaii" #> | `--leaf "Maine" #> `--[dendrogram w/ 2 branches and 38 members at h = 12.5] #> |--[dendrogram w/ 2 branches and 9 members at h = 7.98] #> | |--[dendrogram w/ 2 branches and 7 members at h = 6.12] #> | | |--[dendrogram w/ 2 branches and 4 members at h = 5.03] #> | | | |--leaf "Arizona" #> | | | `--[dendrogram w/ 2 branches and 3 members at h = 4.11] #> | | | |--leaf "Colorado" #> | | | `--[dendrogram w/ 2 branches and 2 members at h = 3.11] #> | | | |--leaf "Montana" #> | | | `--leaf "Nevada" #> | | `--[dendrogram w/ 2 branches and 3 members at h = 3.77] #> | | |--[dendrogram w/ 2 branches and 2 members at h = 2.28] #> | | | |--leaf "Idaho" #> | | | `--leaf "Wyoming" #> | | `--leaf "Utah" #> | `--[dendrogram w/ 2 branches and 2 members at h = 3.06] #> | |--leaf "Kansas" #> | `--leaf "Nebraska" #> `--[dendrogram w/ 2 branches and 29 members at h = 12.5] #> |--[dendrogram w/ 2 branches and 3 members at h = 5.1] #> | |--[dendrogram w/ 2 branches and 2 members at h = 4.49] #> | | |--leaf "Kentucky" #> | | `--leaf "Virginia" #> | `--leaf "Oklahoma" #> `--[dendrogram w/ 2 branches and 26 members at h = 10.4] #> |--[dendrogram w/ 2 branches and 18 members at h = 7.26] #> | |--[dendrogram w/ 2 branches and 14 members at h = 6.79] #> | | |--[dendrogram w/ 2 branches and 3 members at h = 3.72] #> | | | |--leaf "California" #> | | | `--[dendrogram w/ 2 branches and 2 members at h = 2.64] #> | | | |--leaf "Oregon" #> | | | `--leaf "Washington" #> | | `--[dendrogram w/ 2 branches and 11 members at h = 5.82] #> | | |--[dendrogram w/ 2 branches and 9 members at h = 4.93] #> | | | |--[dendrogram w/ 2 branches and 7 members at h = 4.13] #> | | | | |--[dendrogram w/ 2 branches and 2 members at h = 2.97] #> | | | | | |--leaf "Connecticut" #> | | | | | `--leaf "New Hampshire" #> | | | | `--[dendrogram w/ 2 branches and 5 members at h = 3.8] #> | | | | |--[dendrogram w/ 2 branches and 3 members at h = 2.61] #> | | | | | |--[dendrogram w/ 2 branches and 2 members at h = 2.28] #> | | | | | | |--leaf "Illinois" #> | | | | | | `--leaf "New Jersey" #> | | | | | `--leaf "New York" #> | | | | `--[dendrogram w/ 2 branches and 2 members at h = 2.53] #> | | | | |--leaf "Indiana" #> | | | | `--leaf "Ohio" #> | | | `--[dendrogram w/ 2 branches and 2 members at h = 2.77] #> | | | |--leaf "Michigan" #> | | | `--leaf "Pennsylvania" #> | | `--[dendrogram w/ 2 branches and 2 members at h = 3.61] #> | | |--leaf "Delaware" #> | | `--leaf "New Mexico" #> | `--[dendrogram w/ 2 branches and 4 members at h = 4.37] #> | |--[dendrogram w/ 2 branches and 3 members at h = 3.92] #> | | |--[dendrogram w/ 2 branches and 2 members at h = 2.73] #> | | | |--leaf "Iowa" #> | | | `--leaf "South Dakota" #> | | `--leaf "North Dakota" #> | `--leaf "Wisconsin" #> `--[dendrogram w/ 2 branches and 8 members at h = 10.4] #> |--[dendrogram w/ 2 branches and 5 members at h = 6.21] #> | |--leaf "Maryland" #> | `--[dendrogram w/ 2 branches and 4 members at h = 5.71] #> | |--[dendrogram w/ 2 branches and 2 members at h = 3.17] #> | | |--leaf "Missouri" #> | | `--leaf "West Virginia" #> | `--[dendrogram w/ 2 branches and 2 members at h = 3.7] #> | |--leaf "North Carolina" #> | `--leaf "Tennessee" #> `--[dendrogram w/ 2 branches and 3 members at h = 5.53] #> |--[dendrogram w/ 2 branches and 2 members at h = 3.21] #> | |--leaf "Massachusetts" #> | `--leaf "Rhode Island" #> `--leaf "Minnesota"data(agriculture) ## Plot similar to Figure 8 in ref if (FALSE) plot(diana(agriculture), ask = TRUE) plot(diana(agriculture))