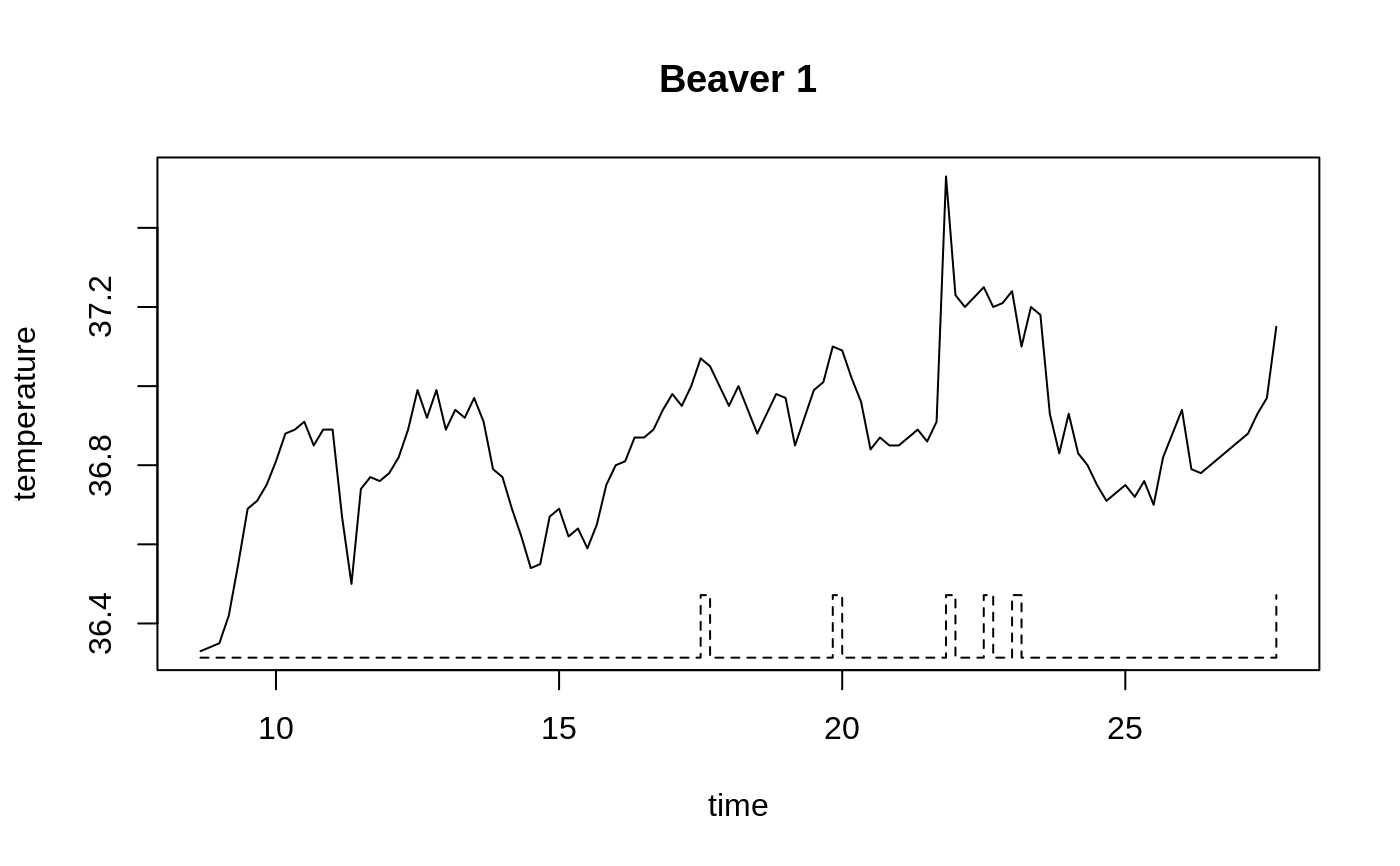
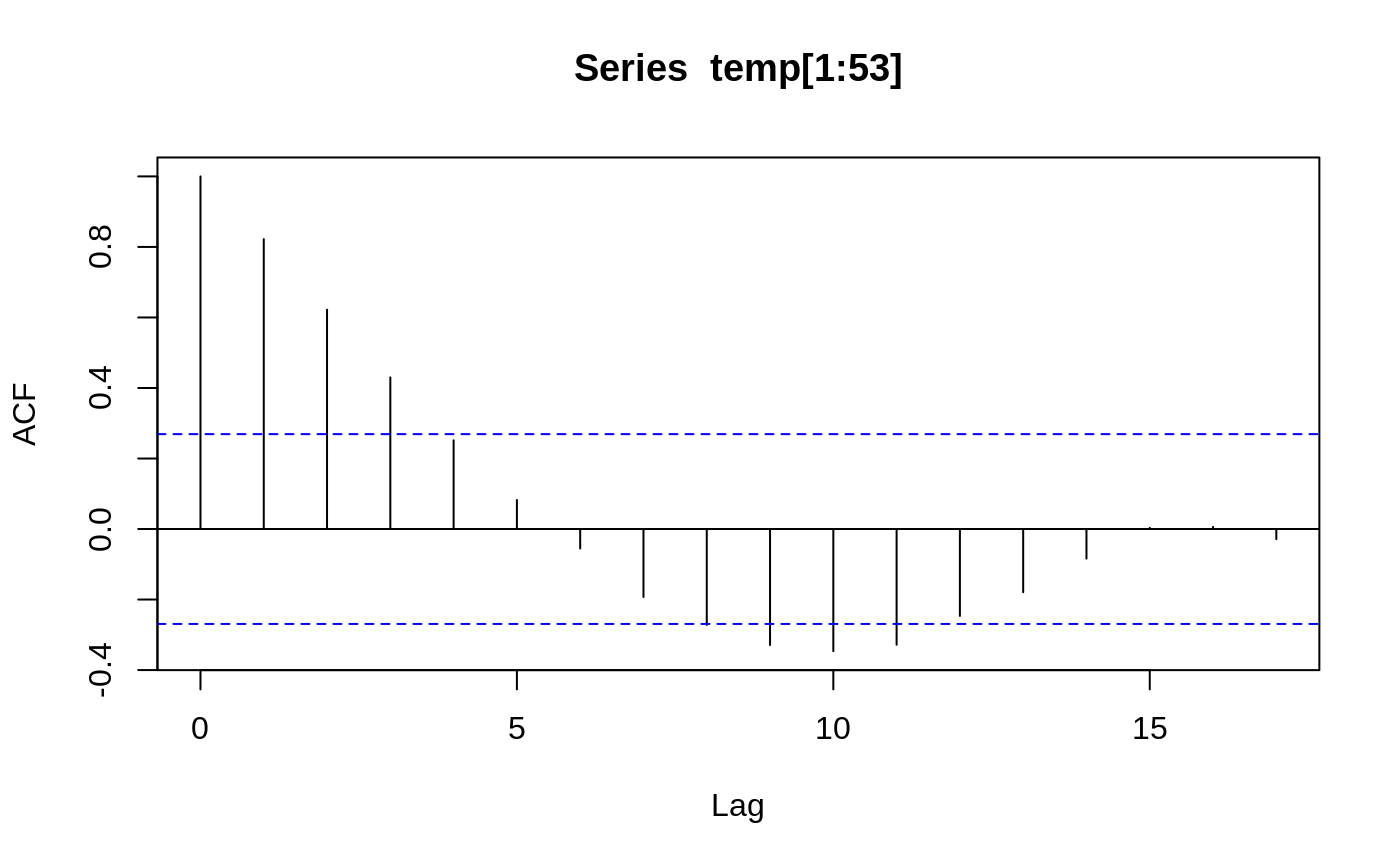
Body Temperature Series of Beaver 1
beav1.RdReynolds (1994) describes a small part of a study of the long-term temperature dynamics of beaver Castor canadensis in north-central Wisconsin. Body temperature was measured by telemetry every 10 minutes for four females, but data from a one period of less than a day for each of two animals is used there.
beav1
Format
The beav1 data frame has 114 rows and 4 columns.
This data frame contains the following columns:
dayDay of observation (in days since the beginning of 1990), December 12--13.
timeTime of observation, in the form
0330for 3.30am.tempMeasured body temperature in degrees Celsius.
activIndicator of activity outside the retreat.
Note
The observation at 22:20 is missing.
Source
P. S. Reynolds (1994) Time-series analyses of beaver body temperatures. Chapter 11 of Lange, N., Ryan, L., Billard, L., Brillinger, D., Conquest, L. and Greenhouse, J. eds (1994) Case Studies in Biometry. New York: John Wiley and Sons.
References
Venables, W. N. and Ripley, B. D. (2002) Modern Applied Statistics with S. Fourth edition. Springer.
See also
Examples
beav1 <- within(beav1, hours <- 24*(day-346) + trunc(time/100) + (time%%100)/60) plot(beav1$hours, beav1$temp, type="l", xlab="time", ylab="temperature", main="Beaver 1")usr <- par("usr"); usr[3:4] <- c(-0.2, 8); par(usr=usr) lines(beav1$hours, beav1$activ, type="s", lty=2)temp <- ts(c(beav1$temp[1:82], NA, beav1$temp[83:114]), start = 9.5, frequency = 6) activ <- ts(c(beav1$activ[1:82], NA, beav1$activ[83:114]), start = 9.5, frequency = 6) acf(temp[1:53])#> #> Call: #> ar(x = temp[1:53]) #> #> Coefficients: #> 1 #> 0.8222 #> #> Order selected 1 sigma^2 estimated as 0.01011act <- c(rep(0, 10), activ) X <- cbind(1, act = act[11:125], act1 = act[10:124], act2 = act[9:123], act3 = act[8:122]) alpha <- 0.80 stemp <- as.vector(temp - alpha*lag(temp, -1)) sX <- X[-1, ] - alpha * X[-115,] beav1.ls <- lm(stemp ~ -1 + sX, na.action = na.omit) summary(beav1.ls, cor = FALSE)#> #> Call: #> lm(formula = stemp ~ -1 + sX, na.action = na.omit) #> #> Residuals: #> Min 1Q Median 3Q Max #> -0.21317 -0.04317 0.00683 0.05483 0.37683 #> #> Coefficients: #> Estimate Std. Error t value Pr(>|t|) #> sX 36.85587 0.03922 939.833 < 2e-16 *** #> sXact 0.25400 0.03930 6.464 3.37e-09 *** #> sXact1 0.17096 0.05100 3.352 0.00112 ** #> sXact2 0.16202 0.05147 3.148 0.00215 ** #> sXact3 0.10548 0.04310 2.448 0.01605 * #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Residual standard error: 0.08096 on 104 degrees of freedom #> (5 observations deleted due to missingness) #> Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999 #> F-statistic: 1.81e+05 on 5 and 104 DF, p-value: < 2.2e-16 #>