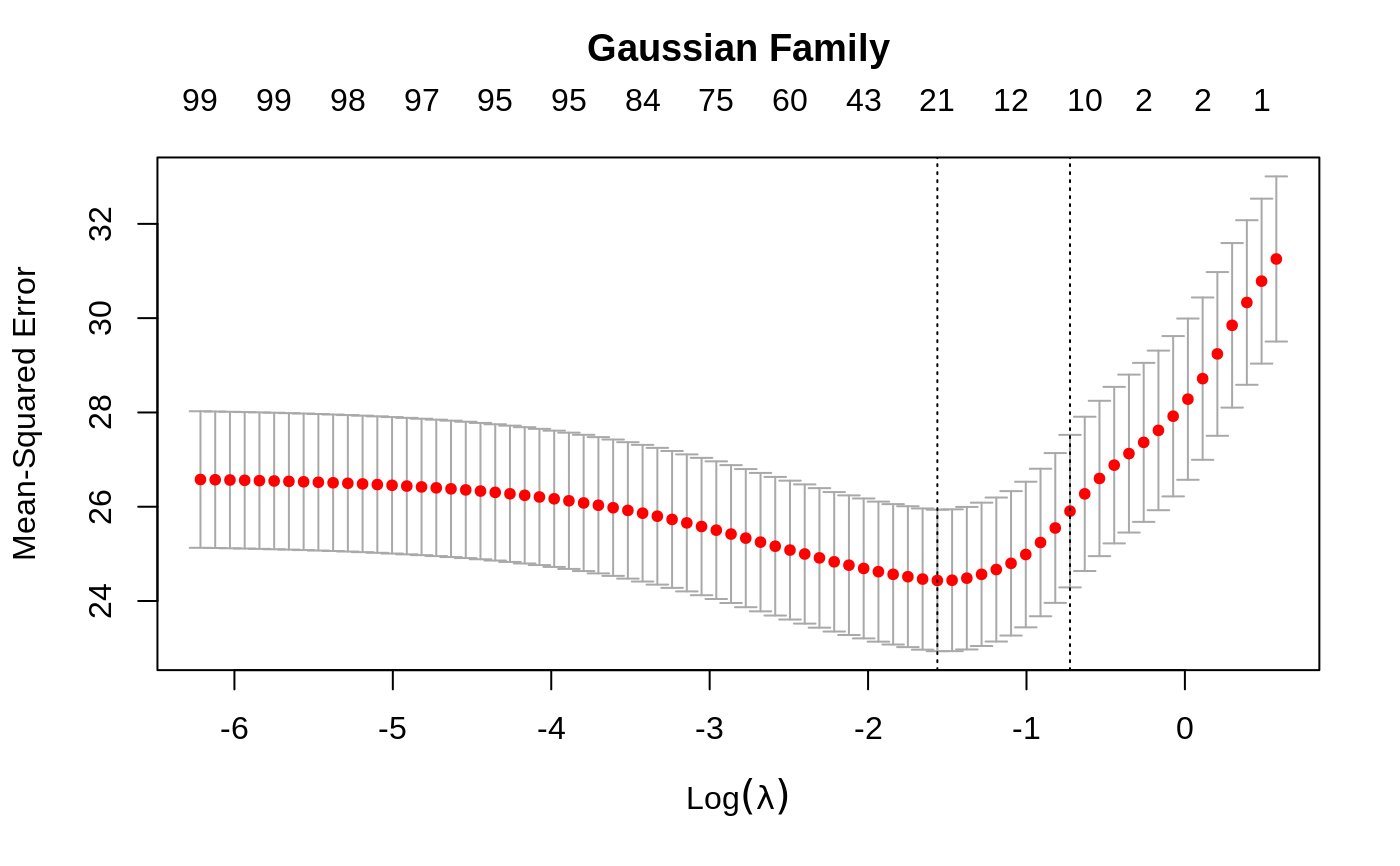
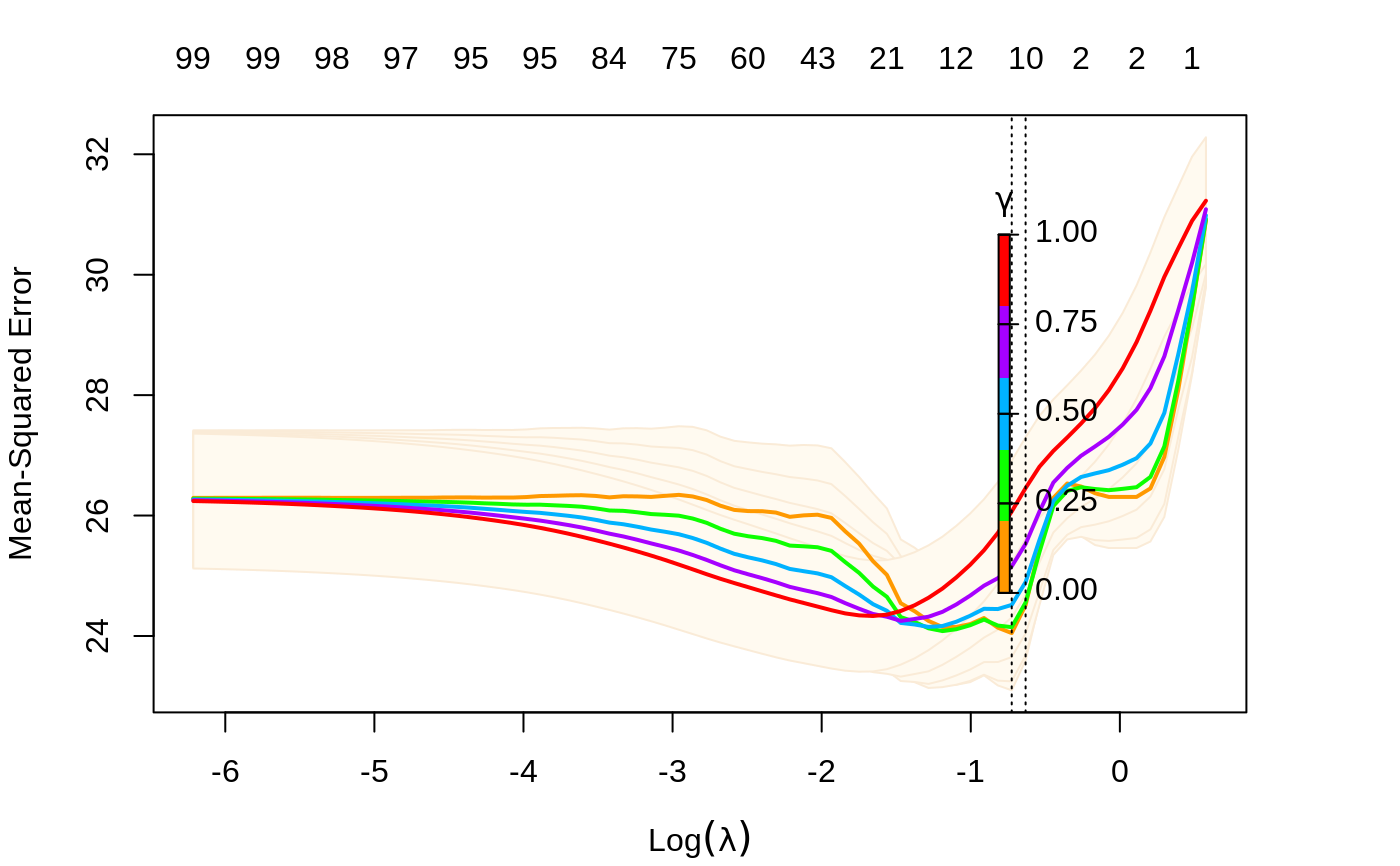
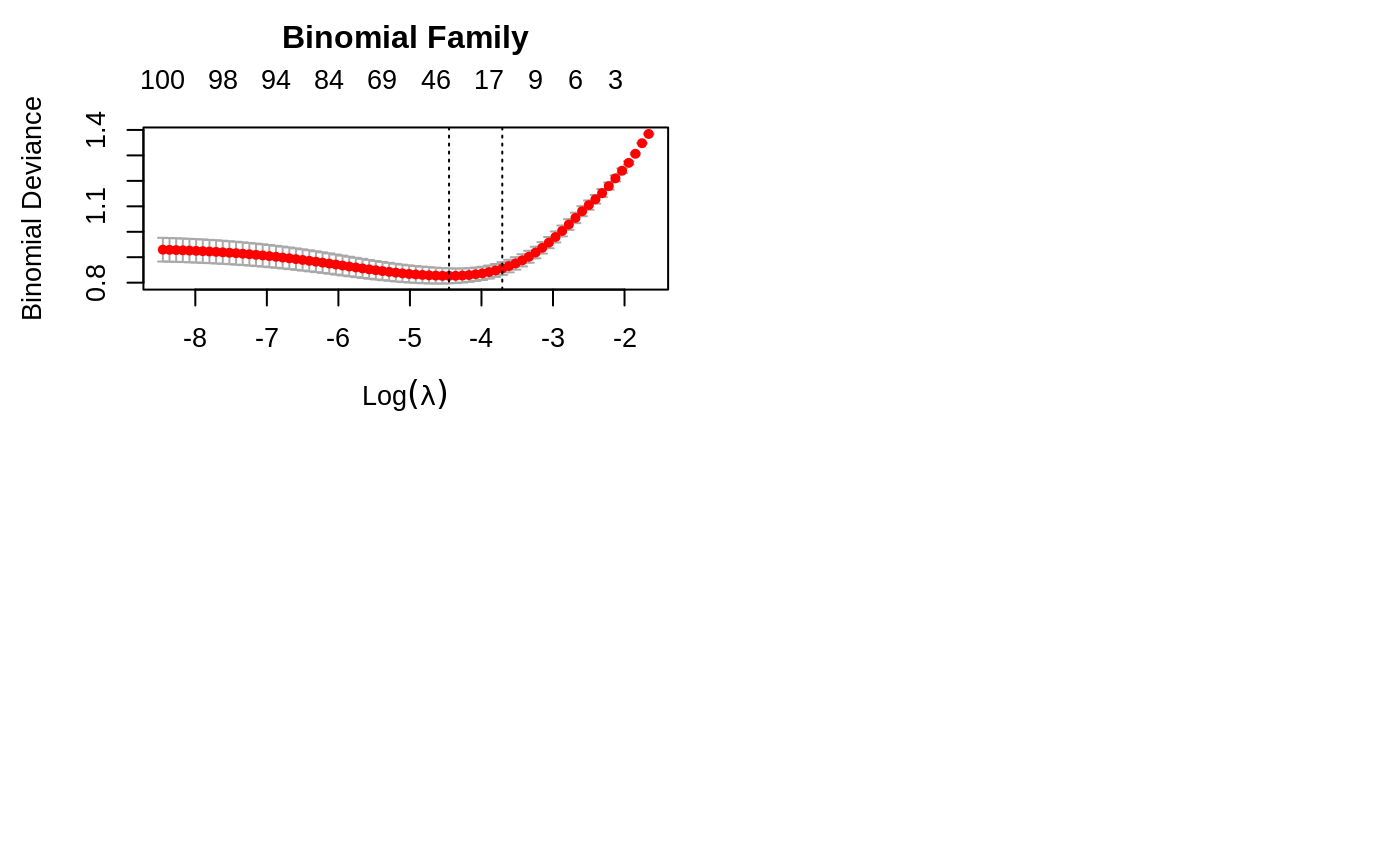
plot the cross-validation curve produced by cv.glmnet
plot.cv.glmnet.RdPlots the cross-validation curve, and upper and lower standard deviation
curves, as a function of the lambda values used. If the object has
class "cv.relaxed" a different plot is produced, showing both
lambda and gamma
# S3 method for cv.glmnet plot(x, sign.lambda = 1, ...) # S3 method for cv.relaxed plot(x, se.bands = TRUE, ...)
Arguments
| x | fitted |
|---|---|
| sign.lambda | Either plot against |
| ... | Other graphical parameters to plot |
| se.bands | Should shading be produced to show standard-error bands;
default is |
Details
A plot is produced, and nothing is returned.
References
Friedman, J., Hastie, T. and Tibshirani, R. (2008) Regularization Paths for Generalized Linear Models via Coordinate Descent
See also
glmnet and cv.glmnet.
Examples
set.seed(1010) n = 1000 p = 100 nzc = trunc(p/10) x = matrix(rnorm(n * p), n, p) beta = rnorm(nzc) fx = (x[, seq(nzc)] %*% beta) eps = rnorm(n) * 5 y = drop(fx + eps) px = exp(fx) px = px/(1 + px) ly = rbinom(n = length(px), prob = px, size = 1) cvob1 = cv.glmnet(x, y) plot(cvob1)frame()set.seed(1011) par(mfrow = c(2, 2), mar = c(4.5, 4.5, 4, 1)) cvob2 = cv.glmnet(x, ly, family = "binomial") plot(cvob2) title("Binomial Family", line = 2.5) ## set.seed(1011) ## cvob3 = cv.glmnet(x, ly, family = "binomial", type = "class") ## plot(cvob3) ## title("Binomial Family", line = 2.5)