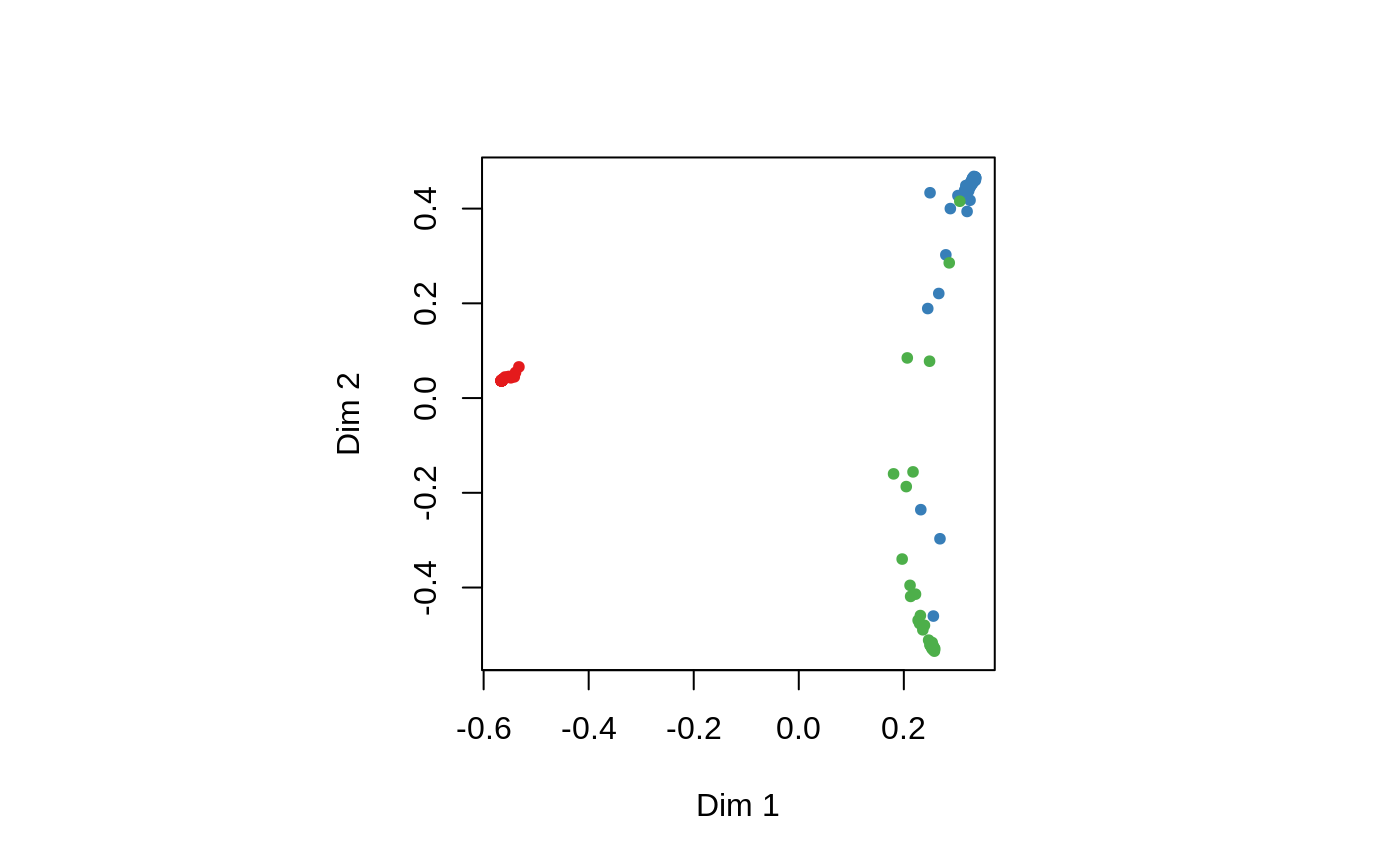
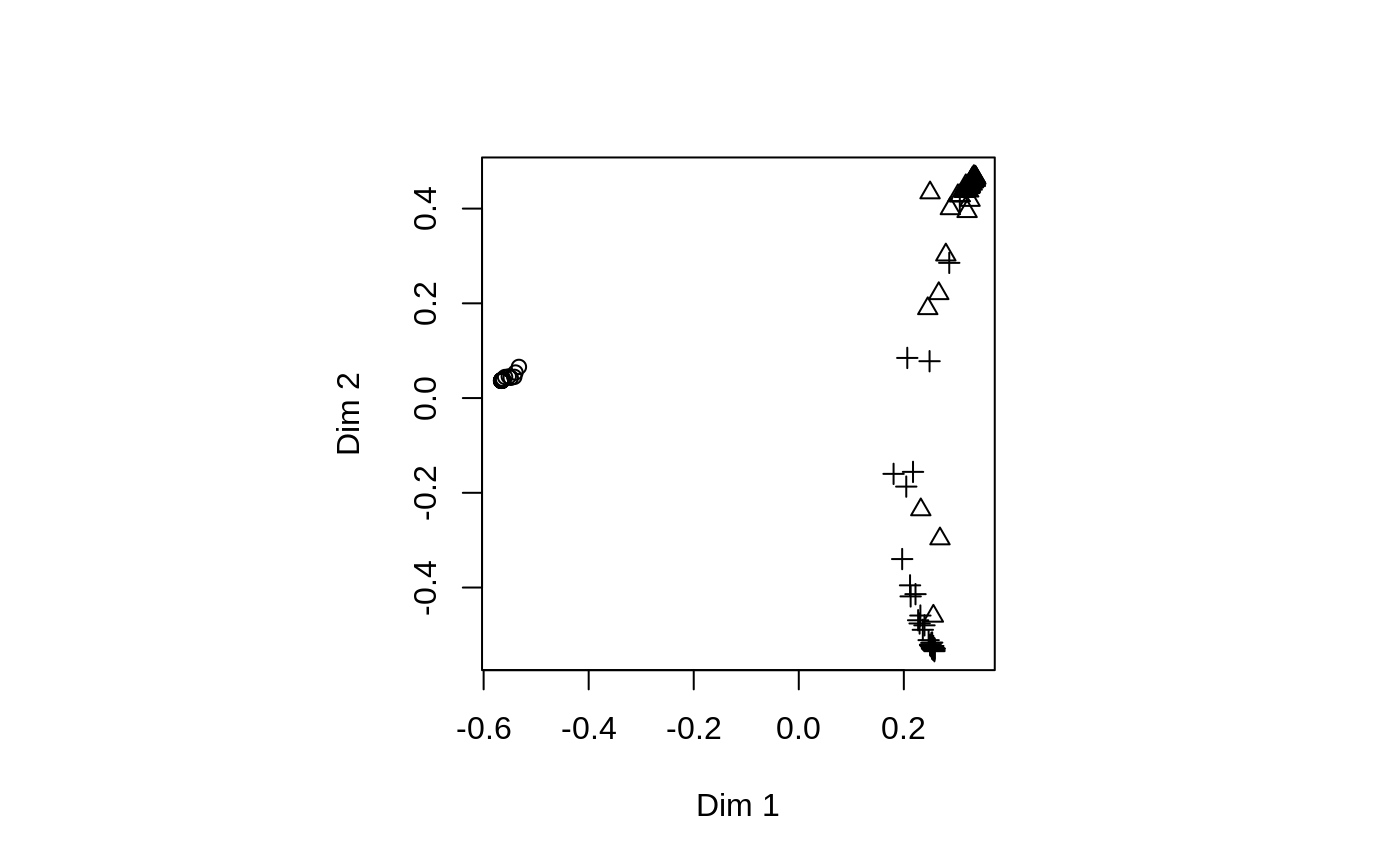
Multi-dimensional Scaling Plot of Proximity matrix from randomForest
MDSplot.RdPlot the scaling coordinates of the proximity matrix from randomForest.
MDSplot(rf, fac, k=2, palette=NULL, pch=20, ...)
Arguments
| rf | an object of class |
|---|---|
| fac | a factor that was used as response to train |
| k | number of dimensions for the scaling coordinates. |
| palette | colors to use to distinguish the classes; length must be the equal to the number of levels. |
| pch | plotting symbols to use. |
| ... | other graphical parameters. |
Value
The output of cmdscale on 1 - rf$proximity is
returned invisibly.
Note
If k > 2, pairs is used to produce the
scatterplot matrix of the coordinates.
See also
Examples
set.seed(1) data(iris) iris.rf <- randomForest(Species ~ ., iris, proximity=TRUE, keep.forest=FALSE) MDSplot(iris.rf, iris$Species)## Using different symbols for the classes: MDSplot(iris.rf, iris$Species, palette=rep(1, 3), pch=as.numeric(iris$Species))