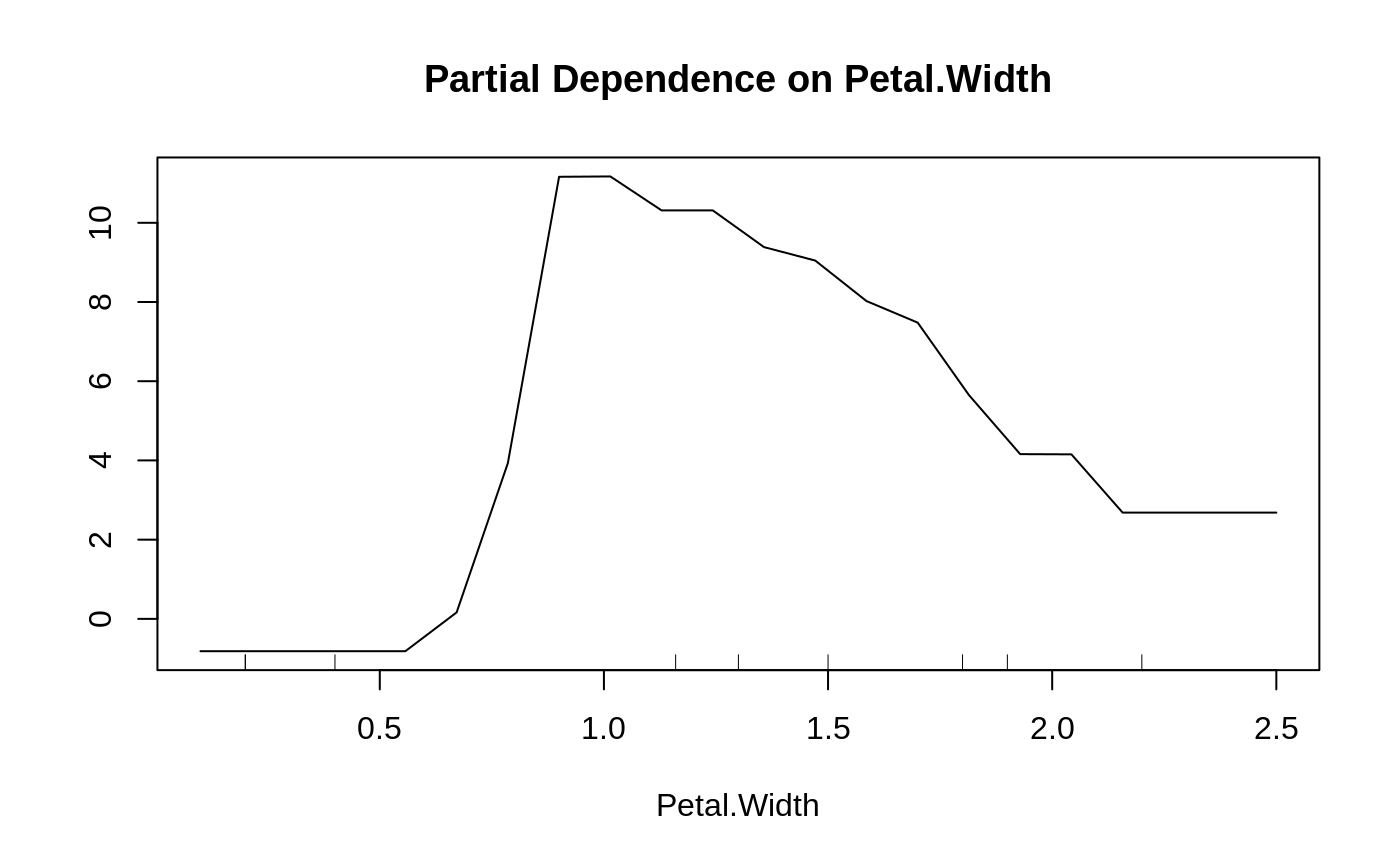
Partial dependence plot
partialPlot.RdPartial dependence plot gives a graphical depiction of the marginal effect of a variable on the class probability (classification) or response (regression).
# S3 method for randomForest partialPlot(x, pred.data, x.var, which.class, w, plot = TRUE, add = FALSE, n.pt = min(length(unique(pred.data[, xname])), 51), rug = TRUE, xlab=deparse(substitute(x.var)), ylab="", main=paste("Partial Dependence on", deparse(substitute(x.var))), ...)
Arguments
| x | an object of class |
|---|---|
| pred.data | a data frame used for contructing the plot, usually the training data used to contruct the random forest. |
| x.var | name of the variable for which partial dependence is to be examined. |
| which.class | For classification data, the class to focus on (default the first class). |
| w | weights to be used in averaging; if not supplied, mean is not weighted |
| plot | whether the plot should be shown on the graphic device. |
| add | whether to add to existing plot ( |
| n.pt | if |
| rug | whether to draw hash marks at the bottom of the plot
indicating the deciles of |
| xlab | label for the x-axis. |
| ylab | label for the y-axis. |
| main | main title for the plot. |
| ... | other graphical parameters to be passed on to |
Value
A list with two components: x and y, which are the values
used in the plot.
Details
The function being plotted is defined as:
$$
\tilde{f}(x) = \frac{1}{n} \sum_{i=1}^n f(x, x_{iC}),
$$
where \(x\) is the variable for which partial dependence is sought,
and \(x_{iC}\) is the other variables in the data. The summand is
the predicted regression function for regression, and logits
(i.e., log of fraction of votes) for which.class for
classification:
$$ f(x) = \log p_k(x) - \frac{1}{K} \sum_{j=1}^K \log p_j(x),$$
where \(K\) is the number of classes, \(k\) is which.class,
and \(p_j\) is the proportion of votes for class \(j\).
Note
The randomForest object must contain the forest
component; i.e., created with randomForest(...,
keep.forest=TRUE).
This function runs quite slow for large data sets.
References
Friedman, J. (2001). Greedy function approximation: the gradient boosting machine, Ann. of Stat.
See also
Examples
data(iris) set.seed(543) iris.rf <- randomForest(Species~., iris) partialPlot(iris.rf, iris, Petal.Width, "versicolor")## Looping over variables ranked by importance: data(airquality) airquality <- na.omit(airquality) set.seed(131) ozone.rf <- randomForest(Ozone ~ ., airquality, importance=TRUE) imp <- importance(ozone.rf) impvar <- rownames(imp)[order(imp[, 1], decreasing=TRUE)] op <- par(mfrow=c(2, 3)) for (i in seq_along(impvar)) { partialPlot(ozone.rf, airquality, impvar[i], xlab=impvar[i], main=paste("Partial Dependence on", impvar[i]), ylim=c(30, 70)) }#> Error in eval(x.var): object 'impvar' not foundpar(op)