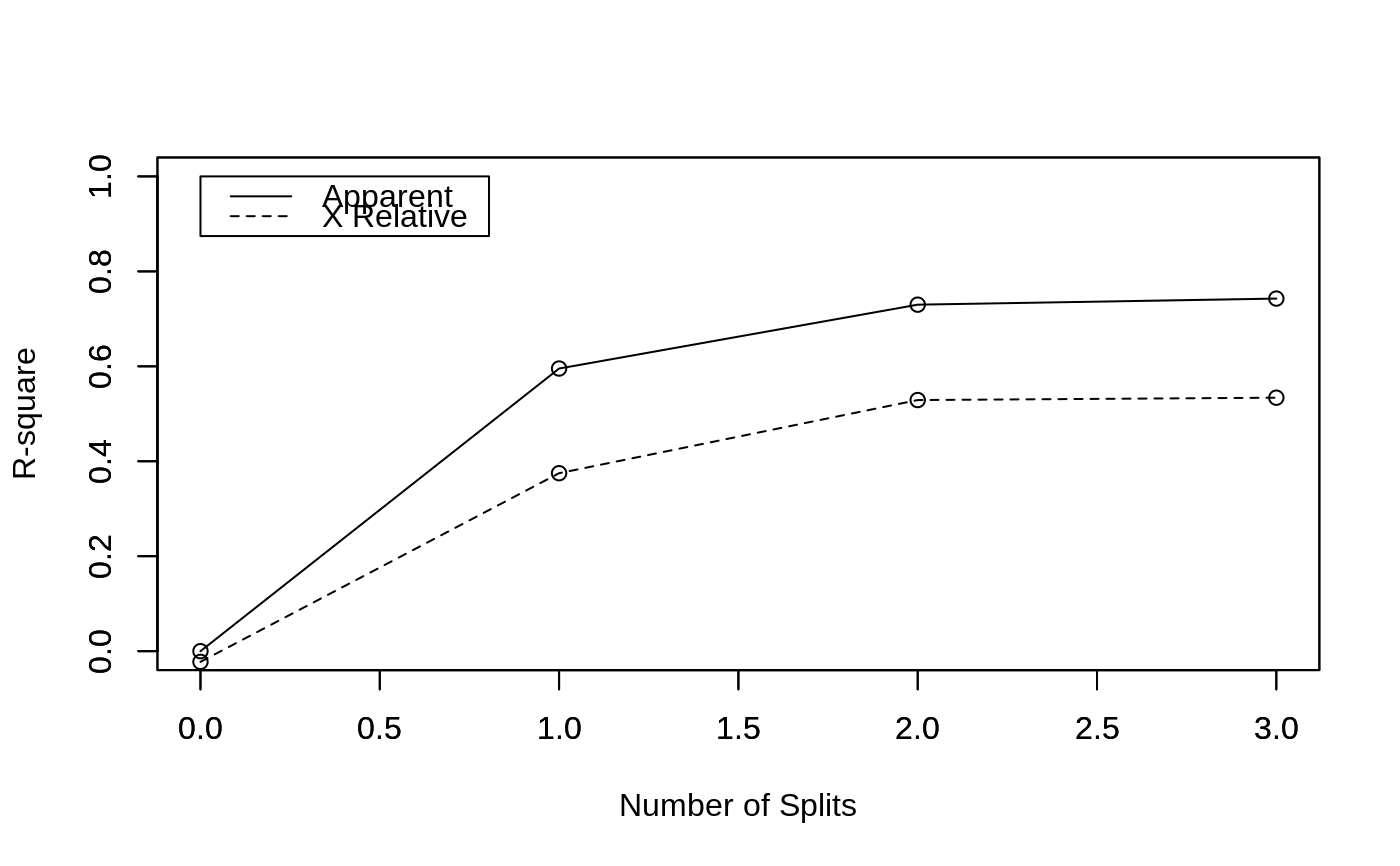
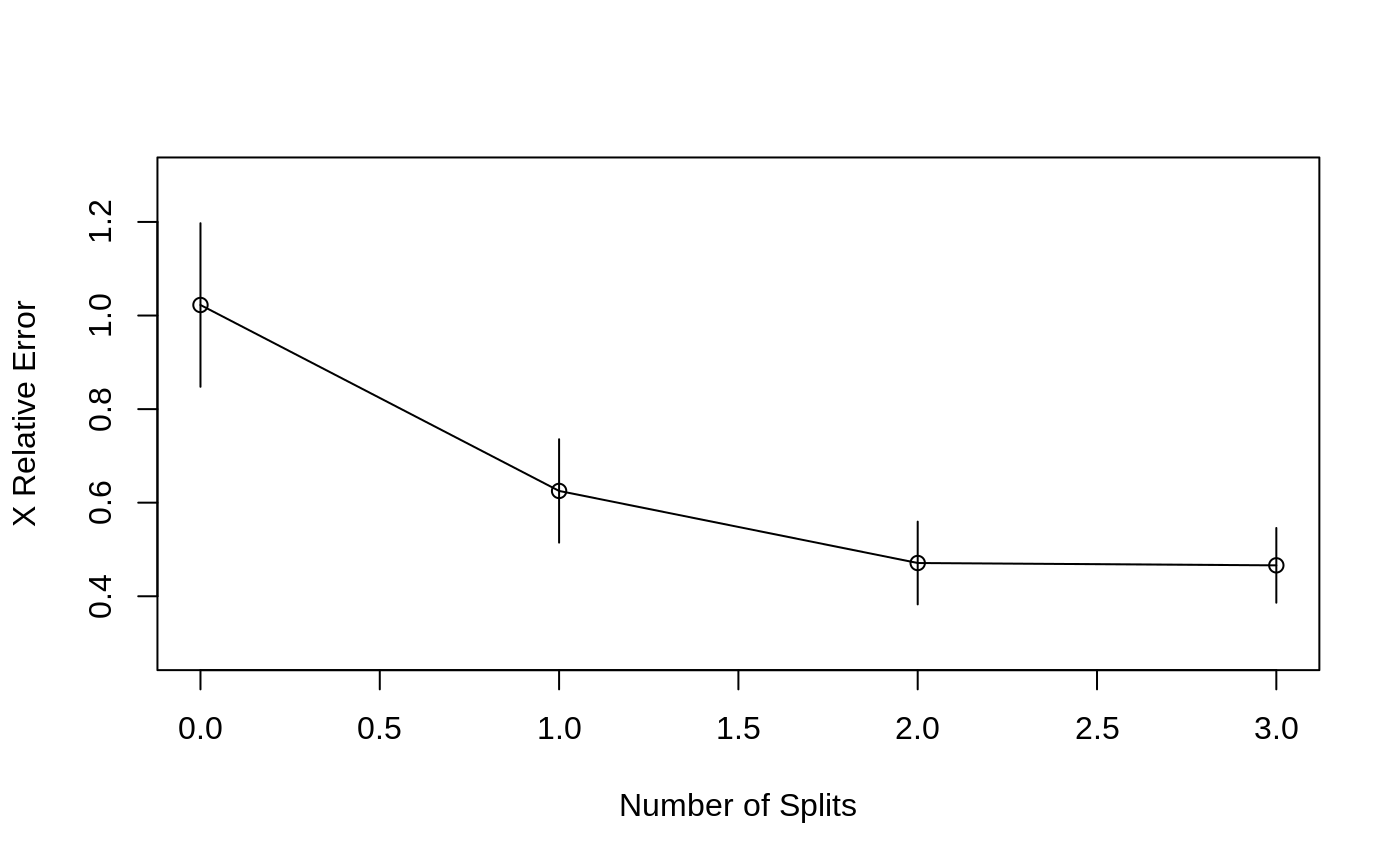
Plots the Approximate R-Square for the Different Splits
rsq.rpart.RdProduces 2 plots. The first plots the r-square (apparent and apparent - from cross-validation) versus the number of splits. The second plots the Relative Error(cross-validation) +/- 1-SE from cross-validation versus the number of splits.
rsq.rpart(x)
Arguments
| x | fitted model object of class |
|---|
Side Effects
Two plots are produced.
Note
The labels are only appropriate for the "anova" method.
Examples
#> #> Regression tree: #> rpart(formula = Mileage ~ Weight, data = car.test.frame) #> #> Variables actually used in tree construction: #> [1] Weight #> #> Root node error: 1354.6/60 = 22.576 #> #> n= 60 #> #> CP nsplit rel error xerror xstd #> 1 0.595349 0 1.00000 1.02253 0.174778 #> 2 0.134528 1 0.40465 0.62512 0.110449 #> 3 0.012828 2 0.27012 0.47106 0.088412 #> 4 0.010000 3 0.25729 0.46608 0.079838